# Pipeline in a Box Containerizing a next generation sequencing pipeline
### In Collaboration With [Elizabeth Bartom](http://www.feinberg.northwestern.edu/faculty-profiles/az/profile.html?xid=32311) [Janna Nugent](http://www.it.northwestern.edu/research/about/rcs-staff.html)
### Ceto Ceto is the name given to the NGS pipeline developed by Elizabeth and her collaborators. [https://github.com/ebartom/NGSbartom/](https://github.com/ebartom/NGSbartom/) 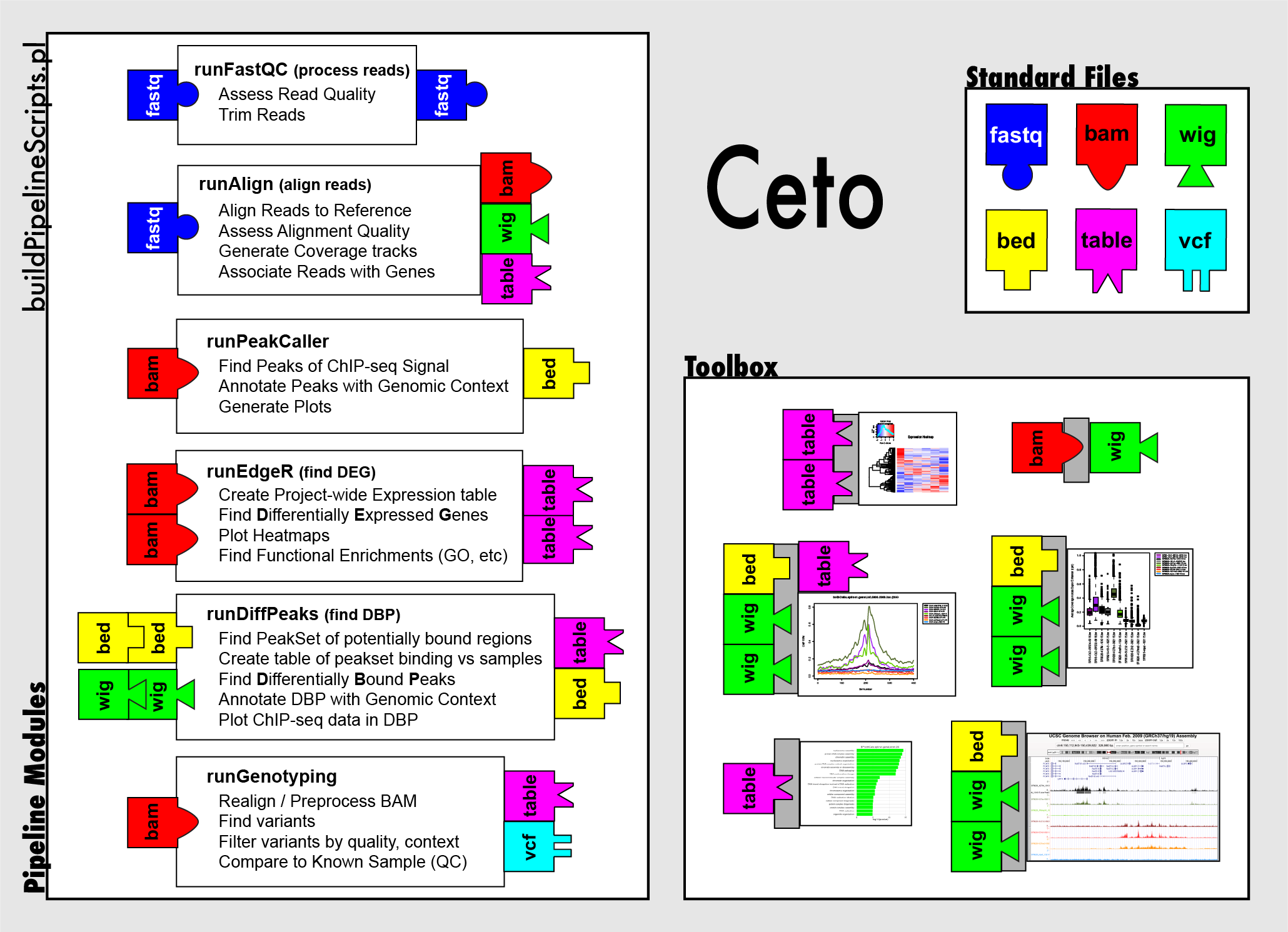 <!-- .element height="50%" width="50%" -->
### Ceto and the Bioinformatics community Elizabeth wished to make Ceto available to the wider bioinformatics community. Namely, people without Quest access.<!-- .element: class="fragment" -->
### Containers are popular for a reason There are benefits to putting code in containers: - easy distribution<!-- .element: class="fragment" --> - easy scaling<!-- .element: class="fragment" --> - encourages good architecture<!-- .element: class="fragment" --> Let's capture those benefits for Ceto!<!-- .element: class="fragment" -->
### Ceto, described A Perl script That writes out multiple shell scripts<!-- .element: class="fragment" --> Which are (optionally) submitted to the Quest scheduler<!-- .element: class="fragment" --> A set of R and Perl scripts, called via the shell scripts, that do post-alignment analysis of samples<!-- .element: class="fragment" --> Documentation<!-- .element: class="fragment" --> (This is how I view it, not how a bioinformatician views it.)<!-- .element: class="fragment" -->
### Ceto, its assumptions - Quest modules<!-- .element: class="fragment" --> - Quest scheduler<!-- .element: class="fragment" --> - Quest storage / filesystem<!-- .element: class="fragment" -->
### Mapping Ceto to Docker Execution environment must match Quest. Must replicate scheduler in some way. "Quest in a box".<!-- .element: class="fragment" -->

### Containerizing Ceto Reading through `buildPipelineScripts.pl` and the documentation to find module dependencies, determine order of execution of shell scripts, and write a wrapper to simulate the scheduler.